A Study of Single Nucleotide Polymorphisms of Tumour Necrosis Factor α-1031 And Tumour Necrosis Factor β+ 252 in Chronic Rhinosinusitis
Article information
Abstract
Objectives
This case-controlled study aimed to identify the association of tumor necrosis factor (TNF)α-1031 and TNFβ+ 252 gene polymorphisms between chronic rhinosinusitis (CRS) and healthy controls. Another purpose of this study was to investigate the associations of these gene polymorphisms with factors related to CRS.
Methods
All deoxyribonucleic acid (DNA) samples were genotyped for TNFα-1031 and TNFβ+252 genes by mean of polymerase chain reaction (PCR) and restriction fragment length polymorphisms (RFLP). The statistical analysis were carried out using chi-square test or Fisher exact test to determine the associations of these gene polymorphisms in CRS. Multiple logistic regression was performed to evaluate the associations of these gene polymorphisms in CRS and its related risk factors.
Results
The genotype and allele frequencies of TNFα-1031 and TNFβ+252 gene did not show any significant associations between CRS and healthy controls. However, a significantly statistical difference of TNFα-1031 was observed in CRS participants with atopy (P-value, 0.045; odds ratio, 3.66) but not in CRS with asthma or aspirin intolerance.
Conclusion
Although the presence of TNFα-1031 and TNFβ+252 gene polymorphisms did not render any significant associations between CRS and healthy control, this study suggests that TNFα-1031 gene polymorphisms in CRS patients with atopy may be associated with increase susceptibility towards CRS.
INTRODUCTION
Chronic rhinosinusitis (CRS) is one of the commonest upper respiratory tract diseases encountered by otorhinolaryngologists as well as primary care physicians. The Global Allergy and Asthma Network of Excellence (GA2LEN) conducted a survey based on European Position Paper on Sinusitis (EPOS) criteria in 19 centres in Europe found that the prevalence of CRS by EPOS criteria was 10.9% [1]. The role of atopy in the development of CRS has been discussed extensively in the literature in which approximately half of the patients with CRS have positive skin prick test [2]. In addition, the concept of ‘one airway, one disease’ proposed the relationship between CRS and asthma [3,4]. Another factors related to CRS is acetylsalicylic acid (ASA) intolerance. Previous study demonstrated 36.0% of patients with aspirin intolerance had concurrent nasal polyps [5].
Mucosal oedema and retention of secretion due to blockage of paranasal sinus ostia is the key to the inflammatory response in CRS. This mucosal irritation of airway epithelium triggers the production of numerous proinflammatory cytokines. Although various cytokines have been proven to play pivotal roles in pathogenesis of CRS, this study concentrated on tumour necrosis factor (TNF). In 1985, Aggarwal et al. [6] was the first who defined 2 distinct types of TNF, which are TNFα and lymphotoxin or also known as TNFβ.
Despite being one of the most prevalent diseases worldwide, the pathogenesis of CRS is still not well understood and appears to be an interaction between environmental and genetic factors. Single nucleotide polymorphisms (SNPs) are defined as genetic variation in deoxyribonucleic acid (DNA) where a single nucleotide is being replaced by another, either by substitution or deletion. The association of SNPs and human disease is complex. Identification of large number of SNPs across human genome has brought the attention of the genetic association studies with common human diseases in a population because common population shares similar genetic variances [7].
The influence of TNF gene polymorphisms in many inflammatory diseases has been addressed in the literatures [8]. The TNF gene polymorphisms have been associated with many chronic inflammatory conditions including rheumatoid arthritis, inflammatory bowel disease and hepatitis B virus infection [9-11]. It has becomes evidence that TNFα antagonists are effective in the treatment for rheumatoid arthritis. Etanercept and Infliximab demonstrated significant improvement of the symptoms in affected patients [12].
Based on growing evidence on the influence of TNF gene polymorphisms in inflammatory diseases, we aimed to determine the association of TNFα-1031 and TNFβ+252 gene polymorphisms between CRS and healthy controls in this study. Besides, we would like to identify the associations of these genes with factors related to CRS.
MATERIALS AND METHODS
Ethical approval
This study had been approved by Human Research Ethics Committee, Universiti Sains Malaysia (Federalwide Assurance Registration No. 00007718; Institutional Review Board No. 00004494).
Sample size calculation
Sample size was determined based on previous literature review by Takeuchi et al. [13] by using PS software ver. 3.1.2, 2014. The power of study was 0.80 with level of statistical significant (α) of 0.05. The exposure among control (healthy participants) was estimated to be 0.56 and exposure among case (CRS participants) was 0.83. The sample size calculation was added with 10% drop out. Therefore, the sample size was 48 participants for cases and 48 participants for controls in order to accept the alternative hypothesis that there is association between TNF gene polymorphisms and CRS and also control.
Case selection
A total of 48 participants with CRS (comprised of 24 chronic rhinosinusitis without nasal polyp [CRSsNP] and 24 chronic rhinosinusitis with nasal polyp [CRSwNP]) were recruited from ORL-HNS Clinic, Hospital Universiti Sains Malaysia following criteria as proposed by European Position Paper on Rhinosinusitis and Nasal Polyps 2012 [14]. A standardised questionnaire determining the demographic characteristics including age, gender, and ethnic origin were obtained and shown in Table 1. Symptoms of CRS which comprised of nasal blockage, rhinorrhea, post nasal drip, facial pain, headache, and loss of smell as well as duration of symptoms were recorded. The presence of asthma, allergy, and ASA intolerance were made primarily by patient’s history. All cases were subjected to general otorhinolaryngologic examination and nasoendoscopic examination. Written consent were taken from all participants.
Control selection
The control groups included 48 healthy volunteers who were not blood-related to the cases were drawn from cases’ friends and spouses. In order to reduce potential environmental bias, the controls were selected from the same district as the cases. A standardised questionnaire determining the demographic characteristics including age, gender, and ethnic origin were taken as depicted in Table 1. They had neither any nasal diseases nor family history of CRS at the time of enrollment. All of them underwent general otorhinolaryngologic examination and had normal nasoendoscopic findings. Written consent were taken from all participants.
DNA extraction
Buccal swab samples were obtained by scrapping the swab firmly against the buccal mucosa and stored at –20°C before DNA extraction was carried out. Extraction of genomic DNA was obtained using Exgene Blood SV Mini (GeneAll, Seoul, Korea).
Polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) for TNFα-1031
The identification of SNPs in the promoter region of TNFα-1031 (rs1799964) was determined using modified protocol as previously described [15]. For amplification of the polymorphic sequence, 0.5 μM forward primer 5´-GGGGAGAACAAAAGGATAAG-3´ and 0.5 μM reverse primer 5´-CCCCATACTCGACTTTCATA-3´ (First BASE Laboratories, Selangor, Malaysia) were mixed together with 5 μL distilled water, 10 μL 2×Phusion PCR Master Mix with High-Fidelity buffer (Thermo Fisher Scientific, Waltham, MA, USA) and 3 μL genomic DNA. The PCR was carried out and the cycling condition included initial denaturation at 98°C for 30 seconds, a total of 30 cycles of denaturation at 98°C for 10 seconds, annealing at 55°C for 30 seconds and extension at 72°C for 10 seconds followed by final extension step at 72°C for 10 minutes. Subsequently, fragment length polymorphism (RFLP) was determined by digestion with restriction enzymes BbsI (New England Biolabs, Hitchin, UK) which cuts the 5´-CCATGG-3´ and 3´-CTTC TG-5´ sequence. A 2.7 μL PCR product were digested with 0.3 μL BbsI restriction enzyme together with 10.5 μL distilled water and 1.5 μL 10×NEBuffer. The mixture was incubated at 37°C for 10 minutes. Seven microlitres of reaction mixture was added to 3 μL 10×BlueJuice Gel Loading buffer (Invitrogen, Carlsbad, CA, USA) and loaded into the gel. The digestion products were separated on 2.5% agarose gel which stained with SYBR Safe DNA gel stain (SBS Genetech, Beijing, China) at 80 V for 50 minutes.
PCR and RFLP for TNFβ+252
The promoter region containing the TNFβ+252 polymorphisms (rs909253) were amplified by PCR using 0.5 μM of 5´-CCGTGCTTCGTGCTTTGGACTA-3´ as forward primer and 5´-AGAGCTGGTGGGACATGTCTG-3´ as reverse primer (SBS Genetech, Beijing, Chin) as described by Takeuchi et al. [13]. The PCR amplification of the TNFβ+252 polymorphisms was carried out in 20 μL volume consisting of 5 μL distilled water, 10 μL 2× Phusion PCR Master Mix with High-Fidelity buffer (Thermo Fisher Scientific), 1 μL forward and reverse primers each and also 3 μL genomic DNA. The cycle parameters consisted of initial denaturation at 98°C for 30 seconds, followed by 30 cycles of denaturation at 98°C for 10 seconds, annealing at 65°C for 20 seconds and extension at 72°C for 5 seconds with a final extension step at 72°C for 10 minutes using Master Cycler Vapo Protect (Eppendorf, Hamburg, Germany). The PCR products underwent NcoI (Thermo Fisher Scientific) digestion which cuts the 5´-CCATGG-3´ and 3´-GGTACC-5´ sequence. A total reaction volume of 30 μL comprised of 17 microlitres of distilled water, 2 μL of 10× FastDigest Green Buffer, 10 μL (~0.2 μg) PCR product and 1 μL FastDigest NcoI enzyme were mixed together and centrifuged briefly. The reaction mixture was then incubated at 37°C for 10 minutes. Ten microliters of this reaction mixture was loaded directly into the gel as the FastDigest Green Buffer can be used as an electrophoresis loading buffer. Electrophoresis of digested product was performed on 2.5% agarose gel which stained with SYBR Safe DNA gel stain at 80 V for 50 minutes.
Statistical analysis
The genotype distributions and allele frequencies was estimated using chi-square test or Fisher exact test to analyse the association between these genepolymorphisms and disease. Multiple Logistic Regression analysis was carried out to determine the associations between these gene polymorphisms and factors related to CRS. All data were entered into IBM SPSS ver. 22.0 (IBM Co., Armonk, NY, USA) for statistical analyses. The results were considered as statistically significant if P-value was less than 0.05. All statistical analyses were two-sided tests.
RESULTS
All 96 samples were successfully genotyped for SNPs for both TNFα-1031 and TNFβ+252 genes. Homozygous wild-type (TT) produced single 270 bp fragment and heterozygous mutant-type (TC) yielded three fragments of 270 bp, 159 bp, and 111 bp as shown in Fig. 1. The homozygous mutant-type (CC) was not detected in both CRS and controls groups. The genotype distributions and allele frequencies for TNFα-1031 are presented in Table 2. The TT genotype was slightly higher in control as compared to CRS participants while the TC genotype was more predominant in CRS. As for allele frequency, T allele appeared slightly higher in control group as compared to CRS but the C allele revealed slightly lower preponderance in control group. However, neither genotype and allele frequencies exhibit significant associations between CRS and healthy controls. Following multivariate analysis, significant association was observed between TNFα-1031 gene polymorphisms and CRS with atopy but lack of associations were seen in CRS with asthma and ASA intolerance as shown in Table 3.
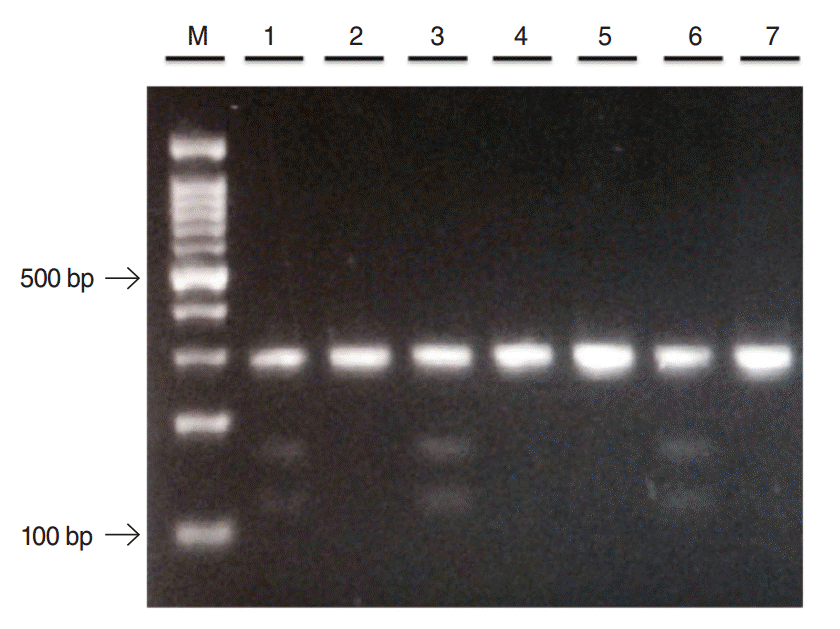
A restriction fragment length polymorphism analysis of tumor necrosis factor-alpha-1031 gene. Lane 2, 4, 5, and 7: homozygous wild-type (TT); lane 1, 3, and 6: heterozygous mutant-type (TC); M: 100 bp DNA ladder.
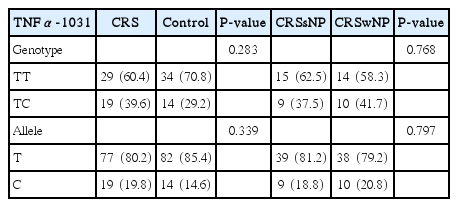
Genotype distributions and allele frequencies of TNFα-1031 among CRS participants and controls as well as CRSsNP and CRSwNP
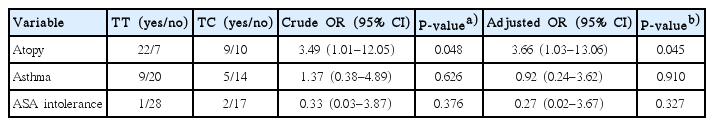
Associations of TNFα-1031 polymorphisms with factors related to CRS adjusted for atopy, asthma and ASA intolerance
Three genotypes of TNFβ+252 polymorphisms were demonstrated in Fig. 2. Homozygous wild-type (AA) produced single fragment of 740 bp, heterozygous mutant-type (AG) yielded 3 fragments of 740 bp, 555 bp, and 185 bp while homozygous mutant-type (GG) showed 2 fragments of 555 bp and 185 bp. The genotype and allele frequencies were homogenously distributed among CRS and control groups. There was no significant difference obtained for TNFβ+252 genotypes distributions and allele frequencies between CRS and control as shown in Table 4. Table 5 demonstrates associations between TNFβ+252 gene polymorphisms and factors related to CRS. However, none of these variables show any significant associations with TNFβ+252 gene polymorphims.

A restriction fragment length polymorphism analysis of tumor necrosis factor-beta+252 gene. Lane 3, 4, and 7: homozygous wildtype (AA); lane 1, 5, and 6: heterozygous mutant-type (AG); lane 2: homozygous mutant-type (GG); M: 100 bp DNA ladder.

Genotype distributions and allele frequencies of TNFβ+252 among CRS participants and controls as well as CRSsNP and CRSwNP
DISCUSSION
In this study, the statistical analysis was unable to demonstrate significant association of TNFα-1031 gene polymorphisms between CRS participants’ genotype distribution in comparison to healthy controls. Similarly, the allele frequency of TNFα-1031 gene did not show any significant associations between CRS and control group. The lack of these associations could be contributed by the fact that SNPs at TNFα polymorphic site are more responsible for genetic markers rather than disease association due to its proximity with polymorphic region of major histocompatibility complex class III at chromosome 6 [16].
In this study, we did not observe any statistically significant difference in genotype distribution as well as allele frequency of TNFα-1031 gene polymorphisms between CRSsNP and CRSwNP despite different inflammatory events and clinical course of these 2 conditions [17]. To date, there is no single study attempted to establish the association between TNFα gene between CRSsNP and CRSwNP. This finding suggests that TNF is part of the inflammatory cascade in both CRSsNP and CRSwNP even though the pathogenesis of these 2 conditions is different.
The most striking observation in our study was the statistically significant difference between TNFα-1031 gene polymorphisms and CRS participants who exhibit atopy after adjusted for asthma and ASA intolerance with P-value of 0.037. This data showed that TT genotype of TNFα-1031 gene increase risk of CRS susceptibility by 3.66-fold in CRS participants with atopy. In contrast, CRS participants with asthma and ASA intolerance were not statistically significant with TNFα-1031 gene polymorphisms.
As for TNFβ+252 gene polymorphisms, there was no statistically significant associations observed between CRS and healthy individuals. Similarly, when we took into consideration the related factors of CRS, which are atopy, asthma and ASA intolerance on multiple logistic regression analyses, none of these variables show significant associations between TNFβ+252 gene polymorphisms with CRS.
As per the literature search, this is the first research to establish the potential role of TNFα-1031 in CRS. To the best of our knowledge, our study provides the first associative evidence of TNFα-1031 gene polymorphisms in CRS patient with atopy in Southeast Asia region. Atopy does modulate more immune response and therefore, CRS patients with atopy have tendency to develop more severe disease manifestation. Literatures have also shown that CRS with atopy associated with more severe computed tomography findings as well as increase need for surgical intervention [18].
Complex and multifactorial processes in the pathogenesis of CRS might contribute these contradicting results. The pathogenesis of CRS is affected by interplay between the immunologic processes, environmental exposure and appropriate management delivered to the patients [19]. Although it is difficult to establish the reason behind these findings, the environmental factor may confound the genetic association of CRS in different geographical areas [20]. The environmental factor in our region which is the tropical rainforest climate might be different from those observed in other studies. It is one of the potential risk factors for atopy and thus, worsens the CRS in atopic patient.
There are few genetic studies that attempted to clarify the role of TNFα gene polymorphisms in the pathogenesis of CRS at other promoter regions in chromosome 6. Several lines of evidence suggested that TNFα-308 and TNFα-238 in the TNFα promoter regions are the SNPs frequently being studied in CRS. Some studies appeared to support the relationship of TNFα gene polymorphisms with CRS [21-23]. However, several other studies could not replicate such associations [13,24,25].
In this study, we could not produce any significant associations of TNFα-1031 gene polymorphisms in CRS participants with asthma and ASA intolerance. This finding is contradicting with previous report in Turkish population where TNFα-308 gene polymorphisms were associated with nasal polyps patients with atopy and ASA intolerance [2]. Another study by Szabo et al. [26] concluded that TNFα-308 gene polymorphisms was a predictive risk for the disease development in CRS patients that have nasal polyps with ASA intolerance. In a more recent study done among Romanian asthmatic patients with nasal polyposis reported that TNFα-857 gene polymorphisms was significantly associated in this group of patients [27].
Although the role of TNFβ+252 gene polymorphism in CRS is not widely studied before, we attempted to establish its role in this case control study. However, this current report is unable to demonstrate any significant difference of genotype and allele frequencies of TNFβ+252 polymorphisms between CRS participants and healthy controls. The lack of association between this gene and CRS is inconsistent with previous findings [13]. It had been shown that genetic diversity among different populations might influence susceptibility to particular diseases in closed ethnic groups [28].
There is lack of association of TNFβ+252 gene polymorphisms between CRSsNP and CRSwNP participants in this study. Various proinflammatory cytokines play roles in inducing chronic inflammation in both CRSsNP and CRSwNP [29]. Therefore, further molecular analysis is warranted since it seems unlikely that any single proinflammatory cytokine would completely explain the susceptibility towards CRS.
To date, there was no research done to determine the association between TNFβ+252 gene polymorphisms and factors related to CRS. Although the influence of TNFβ+252 gene polymorphisms in CRS patients with asthma is still unclear, its role in asthmatic patients has been established previously [30]. Failure to obtain this association might need further stratification of the clinical parameters in larger sample size.
The effects of cytokine gene polymorphisms in the pathogenesis of CRS are still unclear until now. Our result supports the hypothesis that SNPs in proinflammatory cytokines influenced the susceptibility to CRS in our population and would contribute to better understanding of the pathogenesis that underlie CRS. Although TNFα-1031 and TNFβ+252 gene polymorphisms did not confer any genetic susceptibility to CRS as compared to healthy controls, we found that TNFα-1031 gene polymorphisms was significantly associated with CRS participants with atopy. This findings are hoping to produce a change in the current clinical paradigm of CRS. First, we would like to suggest that TNFα-1031 gene might be one of the key players in the pathogenesis of CRS with atopy. Besides, CRS patients with atopy are expected to have clinical benefit from the administration of TNFα antagonist in the future.
In this present study, our data propose that TNFα-1031 and TNFβ+252 gene polymorphisms did not confer any genetic susceptibility to CRS as compared to healthy controls even though the role of TNF as proinflammatory cytokine in CRS is well established in the literature. However, we found that TNFα-1031 gene polymorphisms was significantly associated with CRS participants with atopy. Based on our finding, we would like to draw the possibility of TNFα-1031 gene playing a major role in the pathogenesis of CRS. This result may lead to better understanding of genetic basis of CRS, genetic medicine as well as improving health outcomes for patients in the future.
HIGHLIGHTS
▪ Tumor necrosis factor (TNFα)-1031 and TNFβ+252 gene polymorphisms did not render any significant associations in chronic rhinosinusitis (CRS).
▪ TNFα-1031 gene polymorphisms was significantly associated with CRS participants with atopy.
Notes
No potential conflict of interest relevant to this article was reported.

